Our Research
We study the biophysical mechanisms of gene regulation by quantitatively measuring and modeling sequence-function relationships.
Our experimental work uses massively parallel reporter assays (MPRAs) to measure the effects that variant gene regulatory sequences have on gene expression. We pursue this experimental work in two biological contexts: alternative mRNA splicing in human cells and transcriptional regulation in bacteria.
Our theoretical and computational work develops methods for analyzing the data produced by MPRAs and other highly multiplexed assays. We aim to extract biophysically meaningful models of regulatory sequence function, but also to understand the quantitative nature of sequence-function relationships more broadly. These efforts include deploying robust software for use by the larger genomics community.
Principal Investigator
Justin B. Kinney
Associate Professor and Interim Chair
Simons Center for Quantitative Biology
Cold Spring Harbor Laboratory
Email: jkinney@cshl.edu
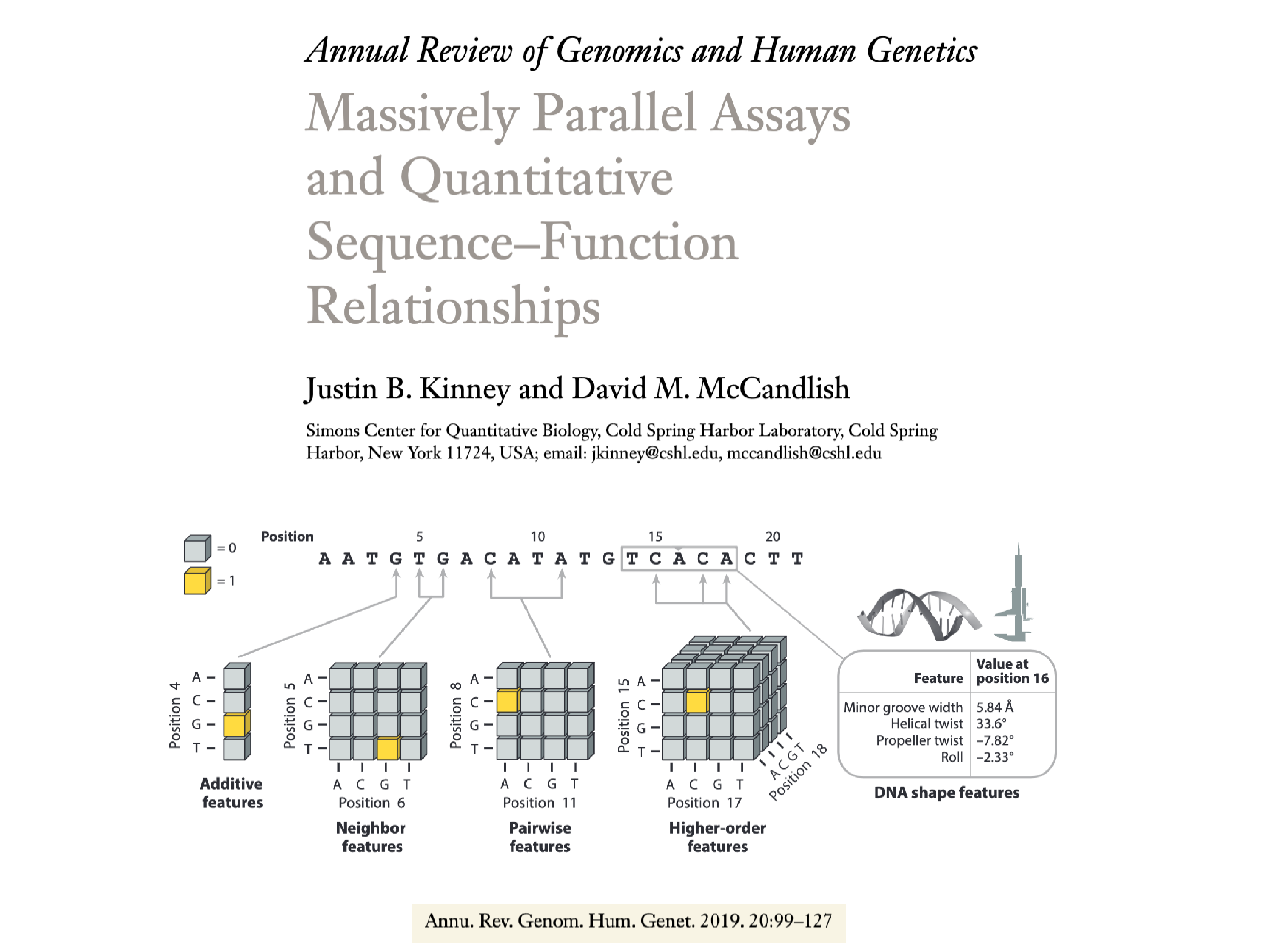
| Kinney JB, McCandlish DM. Massively parallel assays and quantitative sequence-function relationships Annu. Rev. Genomics Hum. Genet. 20:99-127 (2019). |
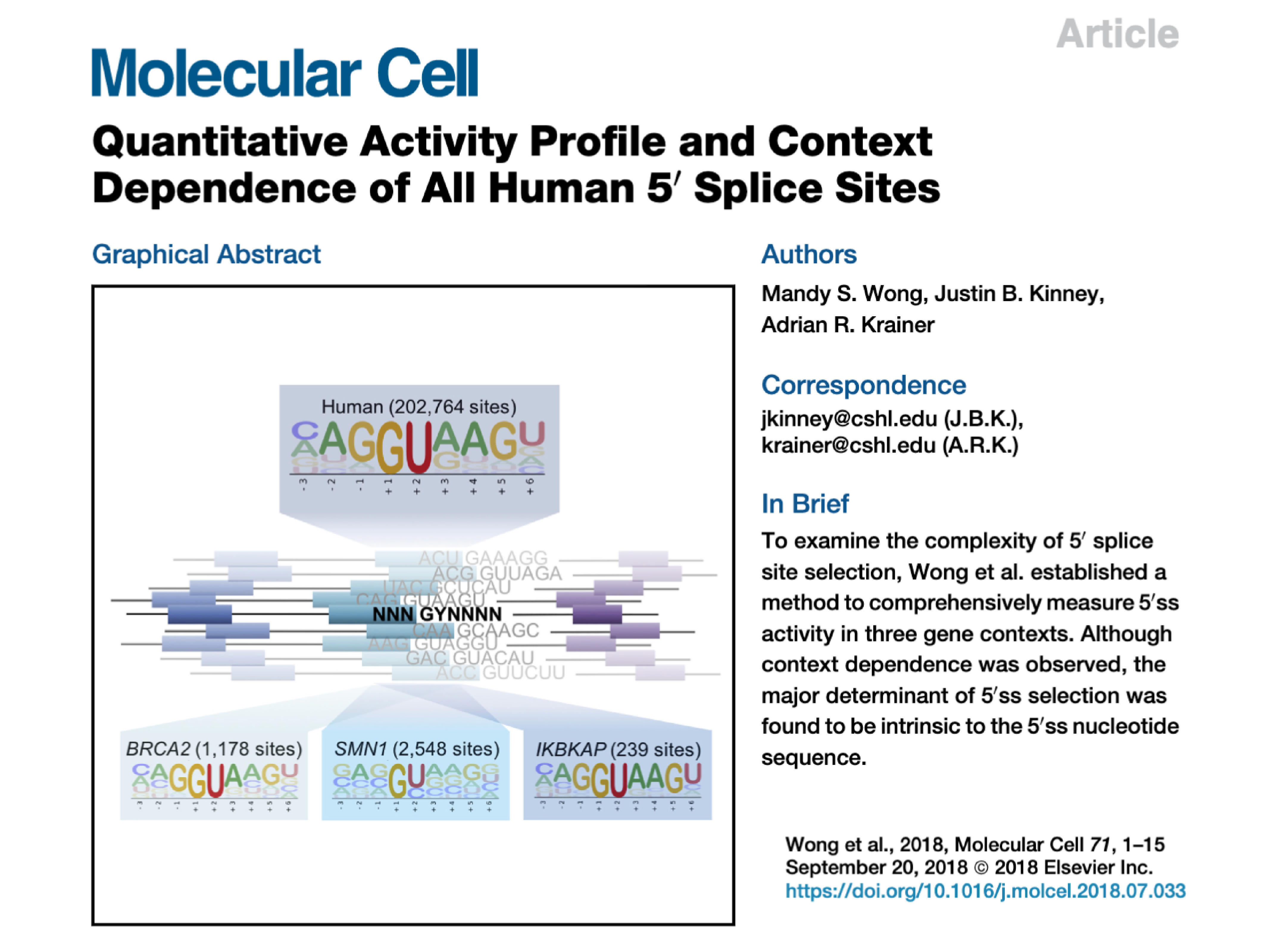
| Wong MS, Kinney JB*, Krainer AR*. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol. Cell 71(6):1012-1026.e3 (2018). *Equal contribution. |
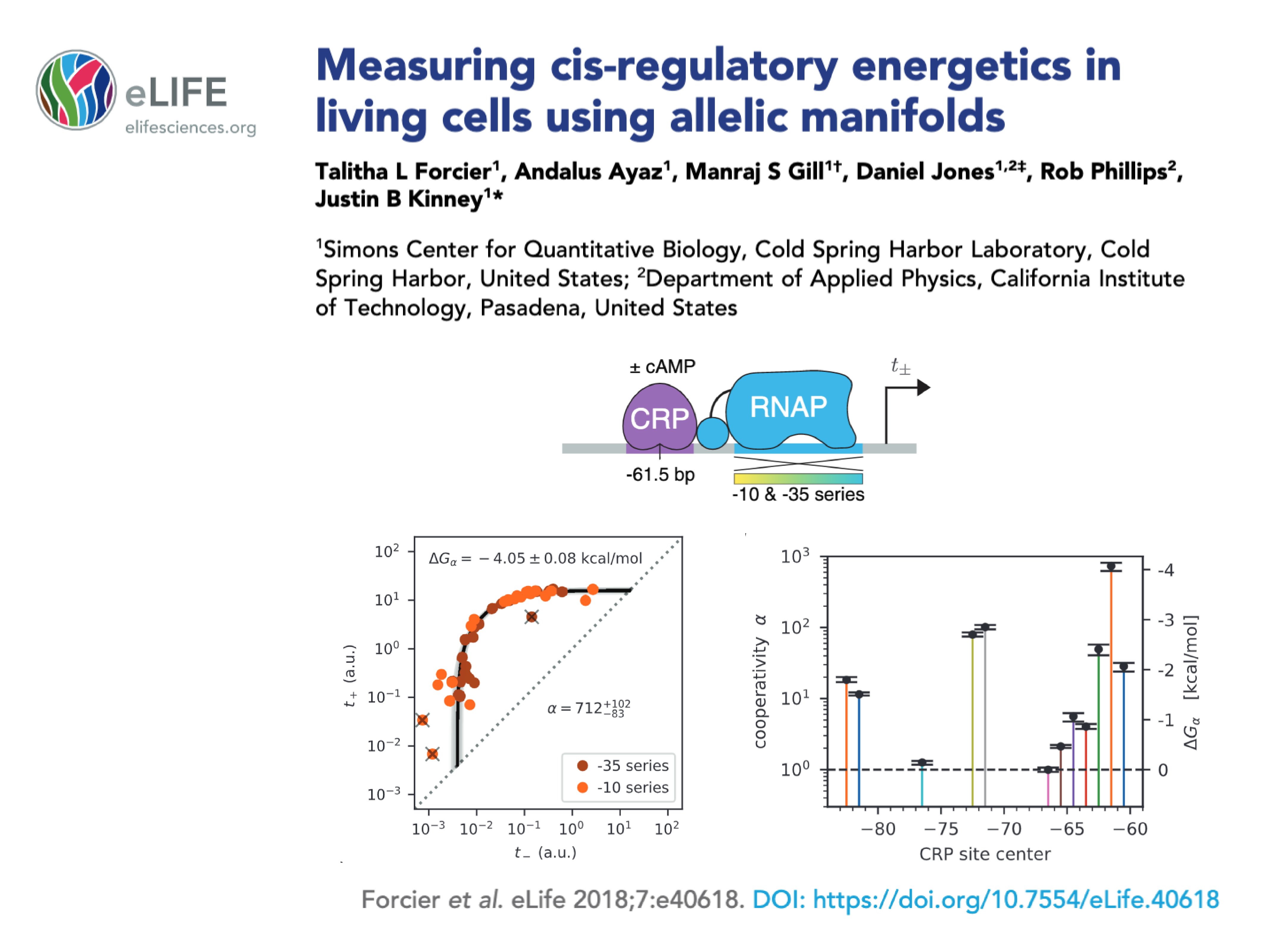
| Forcier T, Ayaz A, Gill MS, Jones D, Phillips R, Kinney JB. Measuring cis-regulatory energetics in living cells using allelic manifolds eLife 7:e40618 (2018). |
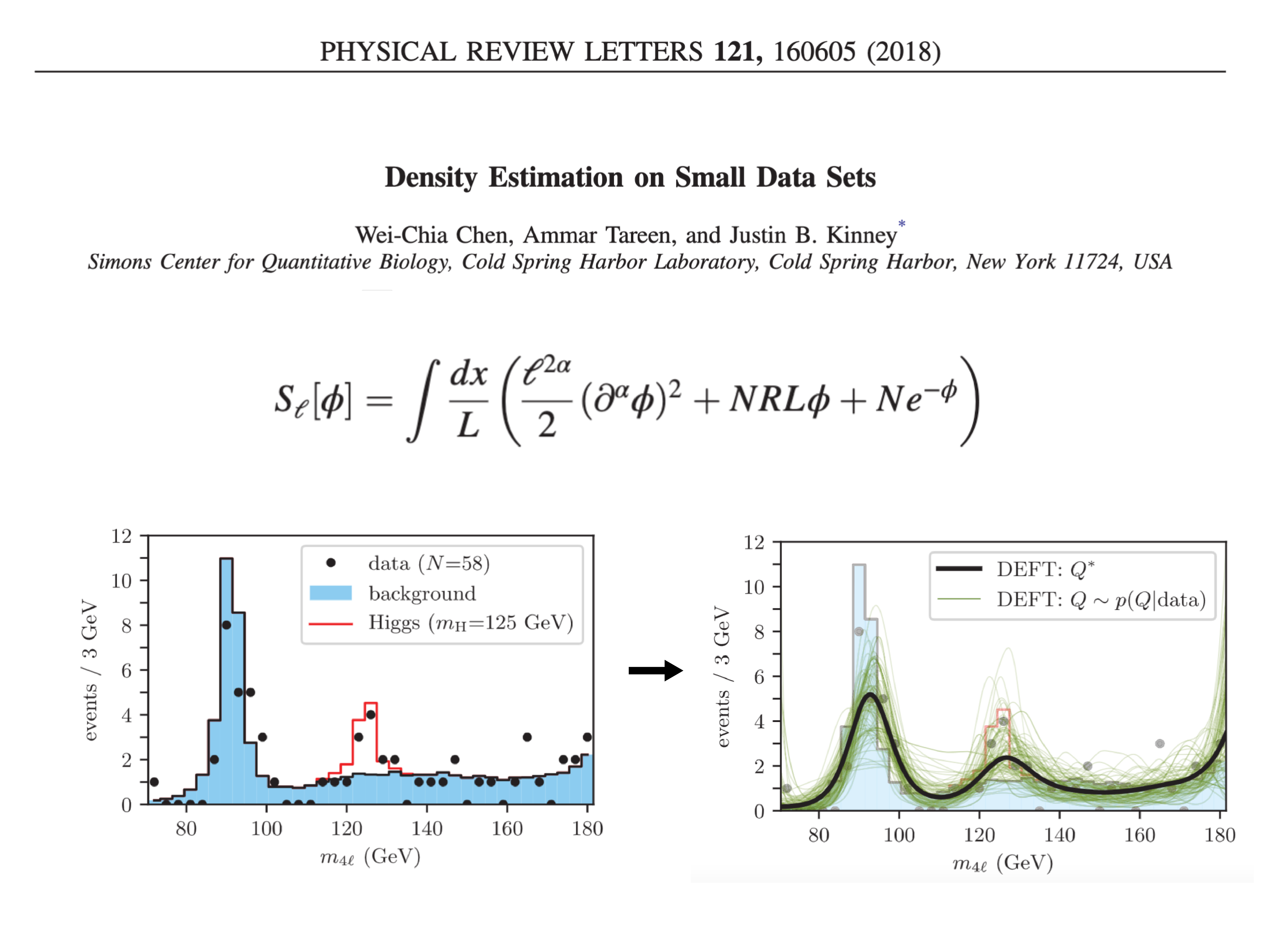
| Chen W, Tareen A, Kinney JB. Density estimation on small datasets. Phys. Rev. Lett. 121,160605 (2018). |
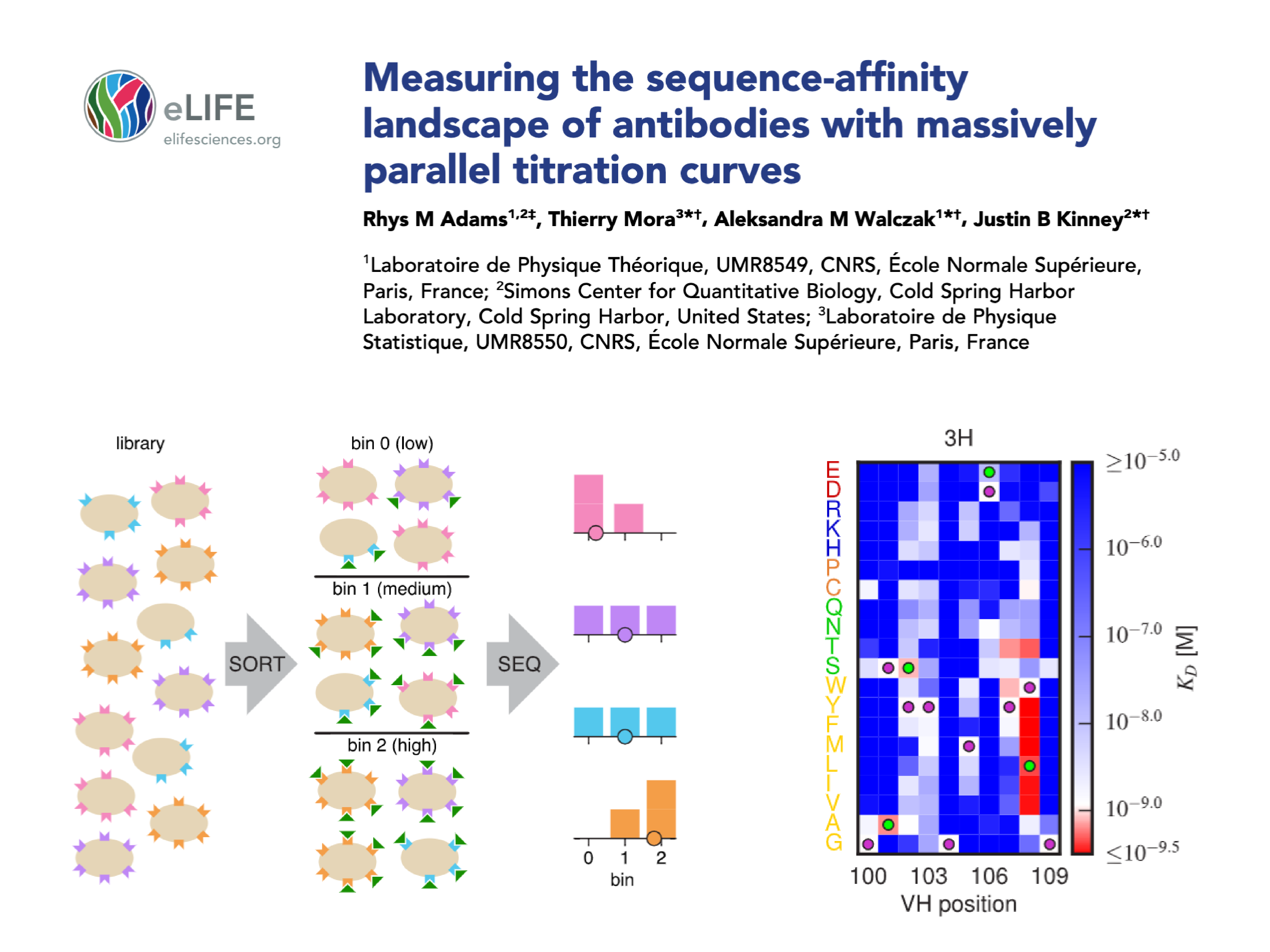
| Adams RM, Mora T*, Walczak AM*, Kinney JB*. Measuring the sequence-affinity landscape of antibodies with massively parallel titration curves. eLife 2016;5:e23156 (2016). *Equal contribution. |
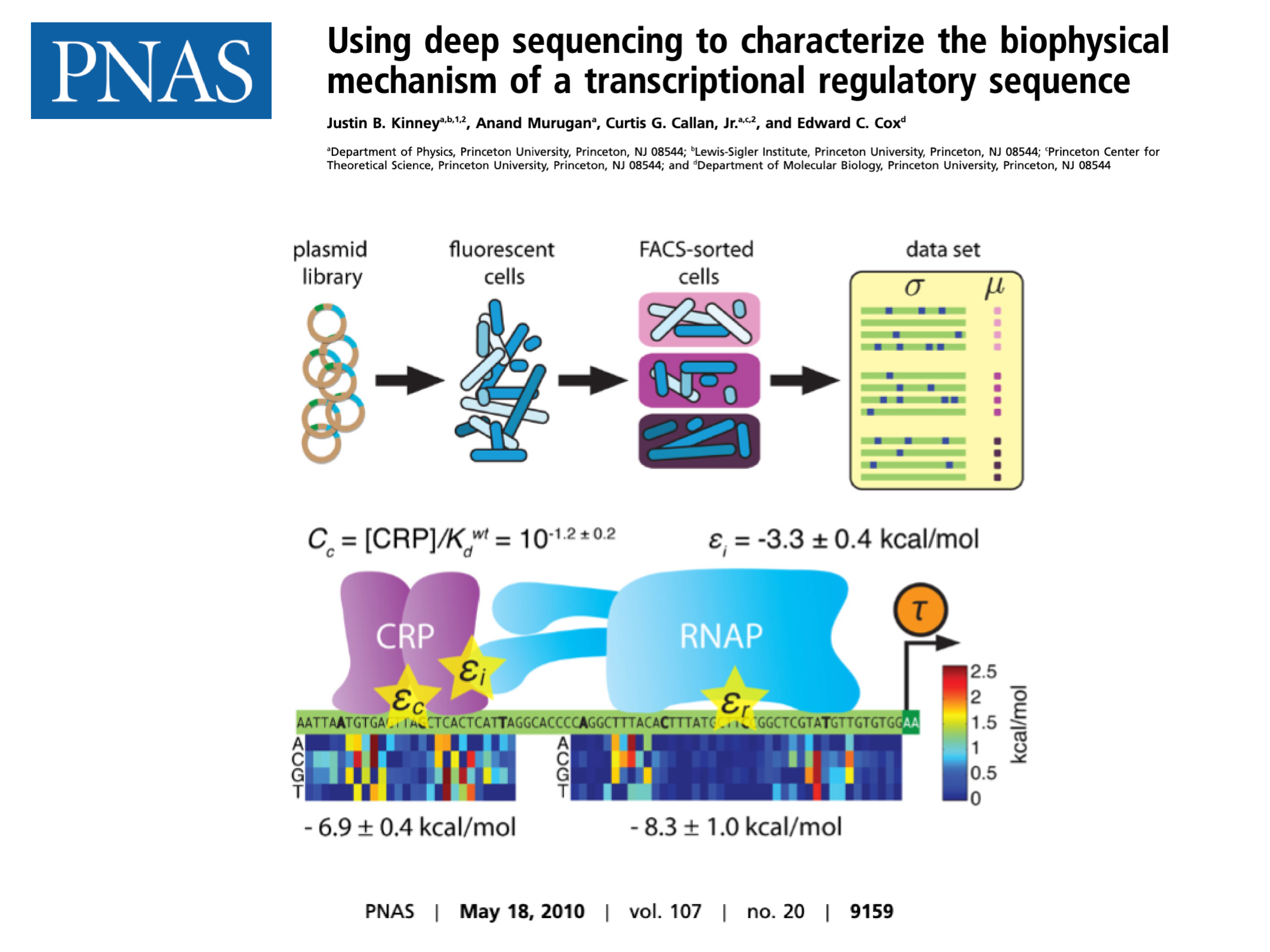
| Kinney JB, Murugan A, Callan CG, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA 107(20):9158-9163 (2010). |